There is a very nice article in the new journal, ACS Central Science
Short Hydrogen Bonds and Proton Delocalization in Green Fluorescent Protein (GFP)
Luke M. Oltrogge and Steven G. Boxer
This is an impressive piece of work spanning from molecular biology to chemistry to quantum physics.
There is also a commentary on the paper by Judith Klinman, placing it in the context of the controversial issue of low-barrier hydrogen bonds in enzymes.
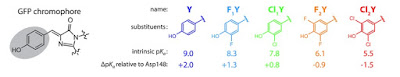
An extensive study was made of mutants of the Green Fluorescent Protein
with a short hydrogen bond between the chromophore and the amino acid Asp148.
The donor-acceptor bond length estimated from
X-ray structures was 2.4 +/- 0.2 Angstroms. This is in the range of low-barrier H-bonds.
What is particularly new here is that through ingenious molecular biology techniques [nonsense suppression] the acidity [pK_a = measure of tendency to give up protons] of the chromophore was systematically varied by 3.5 units
through halogen substitutions.
This range covers the pK_a matching required for strongest H-bonds, as discussed in this earlier post. The experimental results were compared to calculations based
on a one-dimensional proton transfer potential based
on a diabatic state model I have advocated. It was very satisfying for me to see this simple model being used by experimentalists.
To me what is most striking about the paper is the UV absorption spectra below. It is very different from what one normally sees in GFP spectra.
There are generally two absorption bands, denoted A and B, associated with GFP. The A-state and B-state are identified with the neutral chromophore and anionic [i.e. deprotonated] chromophore, respectively. The corresponding spectra are similar to the black and grey curves shown above. The green spectrum above is for the Cl1Y substituted chromophore, which is close to pK_a matching, and is rather broad and intermediate between the A-state and B-state spectra. This is arguably because the proton is delocalised between the chromophore and neighbouring Asp amino acid.
The authors also substituted protons (H) with deuterium (D) to see the extent of quantum nuclear effects. These are normally very small in GFP. However, here they are noticeable.
The measured isotopic fractionation factors Phi (deduced from analysis of the UV absorption spectra)
were in the range
0.54 - 0.9, taking a minimum value for pK_a matching.
This observation and a value of Phi=0.54 for R=2.4 +/- 0.2 Angstroms are consistent with a recent theoretical analysis.
There is one point where I disagree with the theoretical analysis of the authors. I am confused that they average over the vibrational eigenstates to get
an electronic absorption spectrum.
This seems to me this goes against the Franck-Condon principle. If one followed this same procedure for other molecules the UV spectra would
all be much broader than they are, particularly in gas phase.
It is not clear to me how one should proceed in this situation where the proton is quite delocalised and the absorption spectra is significantly different for protonated and de-protonated chromophores. There may be significant Herzberg-Teller effects. One way forward to could be to combine the two-diabatic state H-bond model with a two-state resonance model for the chromophore, such as those advocated by Seth Olsen and I, and then do a full non-adiabatic treatment of the model.
I thank Luke Oltrogge and Seth Olsen for helpful discussions about this work.
Subscribe to:
Post Comments (Atom)
A golden age for precision observational cosmology
Yin-Zhe Ma gave a nice physics colloquium at UQ last week, A Golden Age for Cosmology I learnt a lot. Too often, colloquia are too speciali...

-
This week Nobel Prizes will be announced. I have not done predictions since 2020 . This is a fun exercise. It is also good to reflect on w...
-
Is it something to do with breakdown of the Born-Oppenheimer approximation? In molecular spectroscopy you occasionally hear this term thro...
-
Nitrogen fluoride (NF) seems like a very simple molecule and you would think it would very well understood, particularly as it is small enou...




No comments:
Post a Comment