For a diverse range of chemical compounds, the strength of hydrogen bonds [parametrised by the binding energy and/or bond length] is correlated with a wide range of physical properties such as bond lengths, vibrational frequencies and intensities, and isotope effects. I have posted about many of these and a summary of the main ones is in this paper.
One correlation which is particularly important for practical reasons is the correlation of bond strength (and length) with the chemical shift associated with proton NMR.
The chemical shift is the difference between the NMR resonant frequency of the proton in a specific molecule and that of a free proton. The first important point is that although this shift is extremely small (typically one part in 100,000!) one can measure it extremely accurately.
More importantly, this shift is quite sensitive to the local chemical bonding and so one can use it to actually identify the bonding in unknown molecules (e.g. protein structure determination).
Indeed, if you go to the library and open up a book on NMR or organic chemistry you will find tables and figures giving the chemical shifts associated with different functional groups.
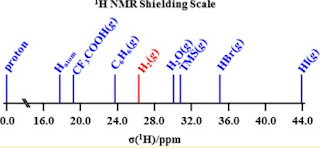
The figure below (taken from here) shows proton chemical shifts for some different molecules.
Why does this happen?
Very roughly the chemical shift is largely determined by the local electronic charge density near the proton and this is modified by the local chemical bonding.
What about hydrogen bonds?
The figure below shows the correlation between the chemical shift and the donor-acceptor distance R for a range of molecules, as found in a 1980 paper.
Confusing aside: the figure shows the chemical shift relative to the standard TMS and so involves negative values.
Why does this matter?
The correlation provides a means to accurately "measure" the donor-acceptor bond length when one does not have direct measurements (e.g. by X-ray diffraction). This is particularly useful in proteins. Indeed, some of the first claims in 1994 on the controversial topic of low-barrier H-bonds (i.e. strong H-bonds) in enzymes were largely based on the observation of unusually small chemical shifts. Some of the subtle issues are discussed here. Another signature is the isotopic fractionation factor.
Although one can calculate these chemical shifts with "black box" computational chemistry, using formulae originally derived by Ramsey in 1950, understanding the underlying physics of the correlations is not clear.
I thank my student Anna Symes for helpful discussions about this topic.
Subscribe to:
Post Comments (Atom)
2025 Nobel Prize in Physics: Macroscopic quantum effects
John Clarke, Michel H. Devoret, and John M. Martinis received the prize “for the discovery of macroscopic quantum mechanical tunnelling an...
-
Is it something to do with breakdown of the Born-Oppenheimer approximation? In molecular spectroscopy you occasionally hear this term thro...
-
This week Nobel Prizes will be announced. I have not done predictions since 2020 . This is a fun exercise. It is also good to reflect on w...
-
Nitrogen fluoride (NF) seems like a very simple molecule and you would think it would very well understood, particularly as it is small enou...




Could you please clarify what "(standard) TMS" means?
ReplyDeleteIt probably is a well-used term since it is not explained in the paper you took the figure from, and googling does not give me a useful direction either.
Thanks
Thanks for the question.
DeleteTMS is a molecule which is used as a reference. All chemical shifts are measured relative to it.
see
https://en.wikipedia.org/wiki/Tetramethylsilane
ah, okay. Thanks for the quick (and simple) answer.
Delete