Here are a few points I came to a better appreciation of:
There are a number of enzymes (e.g. soybean lipoxygenase) which have very small activation energies (Ea~0-2 kcal/mol ~ 100 meV) for hydrogen atom transfers. (n.b. this is a coupled electron and proton transfer). They exhibit kinetic isotope effects which are
- very large in magnitude (~100)
- weakly temperature dependent (difference in Ea for H and D ~ 1 kcal/mol ~ 50 meV)
- change their temperature dependence significantly with mutation
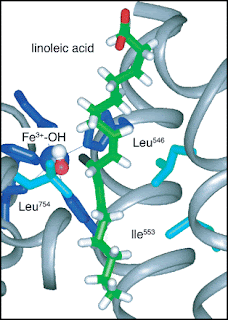
[In the figure above the hydrogen atom (black in the centre of the figure) is transfered to the oxygen atom (red, to the left of the H atom). Mutations correspond to substituting the amino acids Ile553 and/or Leu754.]
The key physics is the following (originally proposed by Kutzenov and Ulstrup) which might be viewed as the proton version on Marcus-Hush electron transfer theory. A JACS paper by Hatcher, Soudakov, and Hammes-Schiffer gives a more sophisticated treatment, including molecular dynamics simulations to extract model parameter values.
The proton directly tunnels between the vibrational ground states of the reactant and product. The isotope effect arises because the spatial extent of the vibrational wavefunction is different for the two isotopes. The temperature dependence of the isotope effect is determined by vibrations of the relative positions of the donor and acceptor atoms.
The main problem or challenge that this model has is the following. The simplest model treatment deduces that the tunneling distance is 0.66 Angstroms [which is less than the van der Waals radii?] and that this increases significantly, up to 2.6 A with mutations. Hammes-Schiffer gets smaller variations, which are more realistic.
However, recent determinations of the crystal structures of the mutants show some structural changes but they do not clearly correlate with the changes in kinetics. In particular there are no detectable changes in tunneling distance. Klinman takes this as evidence for the important role of dynamics.
But, perhaps these structural changes lead to changes in the potential energy surface which in turn changes the amount of tunneling. Things I would like to see include:
-DFT calculations of the potential energy surfaces for the different mutations
-an examination of the Debye-Waller factors for the donor and acceptor atoms in the different mutant structures.
-an examination of non-Born-Oppenheimer effects [which will be isotope dependent].
Note added later: I just found a recent paper by Edwards, Soudakov, and Hammes-Schiffer which uses molecular dynamics to show how the mutations change the tunneling distance and frequency and consequently the kinetic isotope effect. The abstract figure is below.





No comments:
Post a Comment