Proteins are a distinct state of matter. Globular proteins are tightly packed with a density comparable to a crystal but without the spatial regularity found in crystals. The native state is thermodynamically stable, in contrast to the globule state of synthetic polymers which is often glassy and metastable, with a structure that depends on the preparation history.
For a given amino acid sequence the native folded state of the protein is emergent. It has a structure, properties, and function that the individual amino acids do not, nor does the unfolded polymer chain. For example, the enzyme catalase has an active site whose function is as a catalyst to make hydrogen peroxide (which is toxic) decay rapidly.
Protein folding is an example of self-organisation. A key question is how the order of the folded state arises from the disorder (random configuration) of the unfolded state.
There are hierarchies of structures, length scales, and time scales associated with the folding.
The hierarchy of structures are primary, secondary, tertiary, and ternary structures. The primary structure is the amino acid sequence in the heteropolymer. Secondary structures include alpha-helices and beta-sheets, shown in the figure above in orange and blue, respectively. The tertiary structure is the native folded state. An example of a ternary structure is in hemoglobin which consists of four myoglobin units in a particular geometric arrangement.
The hierarchy of time scales varies over more than fourteen orders of magnitude, including folding (msec to sec), helix-coil transitions (microsec), hinge motion (nanosec), and bond vibrations (10 fsecs).
Folding exhibits a hierarchy of processes, summarised in the figure below which is taken from
Masaki Sasai, George Chikenji, Tomoki P. Terada
Modularity
"Protein foldons are segments of a protein that can fold into stable structures independently. They are a key part of the protein folding process, which is the stepwise assembly of a protein's native structure." (from Google AI)
See for example.
Discontinuities
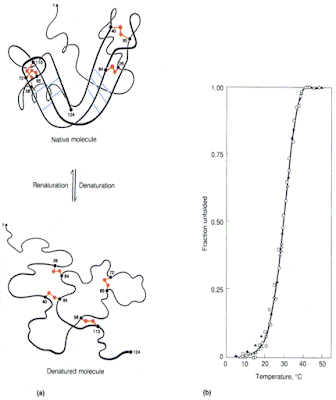
The folding-unfolding transition [denaturation] is a sharp transition, similar to a first-order phase transition. This sharpness reflects the cooperative nature of the transition. There is a well-defined enthalpy and entropy change associated with this transition.
Universality
Proteins exhibit "mutational plasticity", i.e., native structures tolerant to many mutations (changes in individual amino acids). Aspects of the folding process such as its speed, reliability, reversibility, and modularity appear to be universal, i.e., hold for all proteins.
Diversity with limitations
On the one hand, there are a multitude of distinct native structures and associated biological functions. On the other hand, this diversity is much smaller than the configuration space, presumably because thermodynamic stability vasts reduces the options.
Effective interactions
These are subtle. Some of the weak ones matter as the stabilisation energy of the native state is of order 40 kJ per mole, which is quite small as there are about 1000 amino acids in the polymer chain. Important interactions include hydrogen bonding, hydrophobic, and volume exclusion. In the folded state monomers interact with other monomers that are far apart on the chain. The subtle interplay of these competing interactions produces complex structures with small energy differences, as is often the case with emergent phenomena.
Toy models
Note that the interactions are not pairwise but involves strings of "spins" between native contacts.
1. Wako-Saito-Munoz-Eaton model
This is an Ising-like model on a chain. A short and helpful review is
Victor Muñoz
2. Dill's HP polymer on a lattice
This consists of a polymer which has only two types of monomer units and undergoes a self-avoiding walk on a lattice. H and P denote hydrophobic and polar amino acid units, denoted by red and blue circles, respectively, in the figure below. The relative simplicity of the model allows complete enumeration of all possible confirmations for short chains. The model is simpler in two dimensions, yet still captures essential features of the folding problem.
As the H-H attraction increases the chain undergoes a relatively sharp transition to just a few conformations that are compact and have hydrophobic cores. The model exhibits much of the universality of protein folding. Although there are 20 different amino acids in real proteins, the model divides them into two classes and still captures much of the phenomena of folding, including mutational plasticity.
co-operativity - helical order-disorder transition is sharp
Organising principles
Certain novel concepts such as the rugged energy landscape and the folding funnel apply at a particular scale.
This post drew on several nice papers written by Ken Dill and collaborators including
The Protein Folding Problem, H.S. Chan and K.A. Dill, Physics Today, 1993
Roy Nassar, Gregory L. Dignon, Rostam M. Razban, Ken A. Dill, Journal of Molecular Biology, 2021
Interestingly, in the 2021 article, Dill claims that the protein folding problem [which is not the prediction problem] has now essentially been solved.









No comments:
Post a Comment